[R-bloggers] Rmd first: When development starts with documentation (and 7 more aRticles) | |
- Rmd first: When development starts with documentation
- EARLy bird ticket sales end 31 July
- Bang Bang – How to program with dplyr
- R 3.6.1 is now available
- CRAN does not validate R packages!
- Stress Testing Dynamic R/exams Exercises
- Dividend Sleuthing with R
- Tableau – Creating a Waffle Chart
| Rmd first: When development starts with documentation Posted: 10 Jul 2019 05:05 AM PDT (This article was first published on (en) The R Task Force, and kindly contributed to R-bloggers) Documentation matters ! Think about future you and others. Whatever is the aim of your script and analyses, you should think about documentation. The way I see it, R package structure is made for that. Let me try to convince you. At use'R 2019 in Toulouse, I did a presentation entitled: 'The "Rmd first" method: when projects start with documentation'. I ended up saying: Think Package ! If you are really afraid about building a package, you may want to have a look at these slides before. If you are not so afraid, you can start directly with this blog post. In any case,
This article is quite long. You can do a quick read if you skip the bullet points. Use the complete version while you are developing a real project. Why documentation matters ?Prevent code sickness
In any case, think about future you and think about future users or developpers of your work. Never think this is a one shot. One day or another, this one shot script will again be useful. It will be more useful if it is correctly documented.
If you read this post, you are good enough to create a packageYou are not here by chance. You are here because one day, you wrote a line of code in the R language. Thus, trust me, you know enough to create a package. When we tell our students/trainees, after their two first days, that the next step is to build a package, the first reactions are:
Our answers are:
A package forces standardized documentation
There are different places in a package where to write some code, some text, some information, … Remembering the complete procedure of package development may be painful for beginners, even for advanced developers. Hence, some think they need to build a hundred packages before being able to get all of it. That's not true.
Moreover, some useful packages are here to help you in all package development steps, among them {devtools}, {usethis}, {attachment}, {golem}, {desc}, …
Develop as if you were not in a packageIn my presentation at use'R 2019 in Toulouse, I started developing my code in a "classical" R project. If you looked at this presentation, you know we will end up with a package. Here, we will directly start by building a package skeleton. Just a few files to think about before developing. You will see that everything is about documentation. These steps looks like package development…Your project needs to be presented. What is the objective of this work? Who are you? Are others allowed to use your code? We also keep a trace of the history of the package construction steps. Create new "R package using devtools" if you are using Rstudio
Create a file named | |||||||||||||||||||||
| EARLy bird ticket sales end 31 July Posted: 10 Jul 2019 03:21 AM PDT (This article was first published on RBlog – Mango Solutions, and kindly contributed to R-bloggers) EARL London 2019 is getting closer! We've got a great line up of speakers from a huge range of industries and three fantastic keynote speakers – Helen Hunter – Sainsbury's, Julia Silge – Stack Overflow and Tim Paulden – ATASS Sports. Our early bird ticket offer is coming to an end on 31 July, this is your last chance to get EARL tickets at a reduced rate. We hope you can join us for another conference full of inspiration and insights. Also not forgetting our evening networking event, which is the perfect opportunity to catch up with fellow R users over a few (!) free drinks. Check out our highlights for EARL 2018 below and we hope to see you in September! To leave a comment for the author, please follow the link and comment on their blog: RBlog – Mango Solutions. R-bloggers.com offers daily e-mail updates about R news and tutorials on topics such as: Data science, Big Data, R jobs, visualization (ggplot2, Boxplots, maps, animation), programming (RStudio, Sweave, LaTeX, SQL, Eclipse, git, hadoop, Web Scraping) statistics (regression, PCA, time series, trading) and more... This posting includes an audio/video/photo media file: Download Now | |||||||||||||||||||||
| Bang Bang – How to program with dplyr Posted: 10 Jul 2019 03:00 AM PDT (This article was first published on r-bloggers – STATWORX, and kindly contributed to R-bloggers) The tidyverse is making the life of a data scientist a lot easier. That's why we at STATWORX love to execute our analytics and data science with the tidyverse. Its user-centered approach has many advantages. Instead of the base R version Our new function uses Non-Standard Evaluation
The quoting used by  Because dplyr quotes its arguments, we have to do two things to use it in our function:
We will see this quote-and-unquote pattern consequently through functions which are using tidy evaluation. Therefore, as input in our function, we quote the If we are not using Data Mask Generally speaking, the data mask approach is much more convenient. However, on the programming site, we have to pay attention to some things, like the quote-and-unquote pattern from before. As a next step, we want the quotation to take place inside of the function, so the user of the function does not have to do it. Sadly, using We are getting an error message because All R code is a treeTo better understand what's happening, it is useful to know that every R code can be represented by an Abstract Syntax Tree (AST) because the structure of the code is strictly hierarchical. The leaves of an AST are either symbols or constants. The more complex a function call gets, the deeper an AST is getting with more and more levels. Symbols are drawn in dark-blue and have rounded corners, whereby constants have green borders and square corners. The strings are surrounded by quotes so that they won't be confused with symbols. The branches are function calls and are depicted as orange rectangles.  To understand how an expression is represented as an AST, it helps to write it in its prefix form.  There is also the R package called lobstr, which contains the function The code from the first example  It looks as expected and just like our hand-drawn AST. The concept of ASTs helps us to understand what is happening in our function. So, if we have the following simple function, !!` introduces a placeholder (promise) for x. Due to R's lazy evaluation, the function  Furthermore, Perfecting our functionNow, we want to add an argument for the variable we are summarizing to refine our function. At the moment we have Firstly, Tidy DotsIn the next step, we want to add the possibility to summarize an arbitrary number of variables. Therefore, we need to use tidy dots (or dot-dot-dot)
In Within the function,  With using purrr, we can neatly handle the computation with our list entries provided by And the tidy evaluation goes on and onAs mentioned in the beginning, tidy evaluation is not only used within dplyr but within most of the packages in the tidyverse. Thus, to know how tidy evaluation works is also helpful if one wants to use ggplot in order to create a function for a styled version of a grouped scatter plot. In this example, the function takes the data, the values for the x and y-axes as well as the grouping variable as inputs:  Another example would be to use R Shiny Inputs in a sparklyr-Pipeline. server.R ui.R ConclusionThere are many use cases for tidy evaluation, especially for advanced programmers. With the tidyverse getting bigger by the day, knowing tidy evaluation gets more and more useful. For getting more information about the metaprogramming in R and other advanced topics, I can recommend the book Advanced R by Hadley Wickham. ABOUT USSTATWORX
Sign Up Now!
Sign Up Now!
Der Beitrag Bang Bang – How to program with dplyr erschien zuerst auf STATWORX. To leave a comment for the author, please follow the link and comment on their blog: r-bloggers – STATWORX. R-bloggers.com offers daily e-mail updates about R news and tutorials on topics such as: Data science, Big Data, R jobs, visualization (ggplot2, Boxplots, maps, animation), programming (RStudio, Sweave, LaTeX, SQL, Eclipse, git, hadoop, Web Scraping) statistics (regression, PCA, time series, trading) and more... This posting includes an audio/video/photo media file: Download Now | |||||||||||||||||||||
| Posted: 10 Jul 2019 02:42 AM PDT (This article was first published on Revolutions, and kindly contributed to R-bloggers) On July 5, the R Core Group released the source code for the latest update to R, R 3.6.1, and binaries are now available to download for Windows, Linux and Mac from your local CRAN mirror. R 3.6.1 is a minor update to R that fixes a few bugs. As usual with a minor release, this version is backwards-compatible with R 3.6.0 and remains compatible with your installed packages. Nonetheless, an upgrade is recommended to address issues including:
See the release notes for the complete list. By long tradition the code name for this release, "Action of the Toes", is likely a reference to the Peanuts comic. If you know specifically which one, let us know in the comments! R-announce mailing list: R 3.6.1 is released To leave a comment for the author, please follow the link and comment on their blog: Revolutions. R-bloggers.com offers daily e-mail updates about R news and tutorials on topics such as: Data science, Big Data, R jobs, visualization (ggplot2, Boxplots, maps, animation), programming (RStudio, Sweave, LaTeX, SQL, Eclipse, git, hadoop, Web Scraping) statistics (regression, PCA, time series, trading) and more... This posting includes an audio/video/photo media file: Download Now | |||||||||||||||||||||
| CRAN does not validate R packages! Posted: 09 Jul 2019 03:19 PM PDT (This article was first published on R – Xi'an's Og, and kindly contributed to R-bloggers)
To leave a comment for the author, please follow the link and comment on their blog: R – Xi'an's Og. R-bloggers.com offers daily e-mail updates about R news and tutorials on topics such as: Data science, Big Data, R jobs, visualization (ggplot2, Boxplots, maps, animation), programming (RStudio, Sweave, LaTeX, SQL, Eclipse, git, hadoop, Web Scraping) statistics (regression, PCA, time series, trading) and more... | |||||||||||||||||||||
| Stress Testing Dynamic R/exams Exercises Posted: 09 Jul 2019 03:00 PM PDT (This article was first published on R/exams, and kindly contributed to R-bloggers) Before actually using a dynamic exercise in a course it should be thoroughly tested. While certain aspects require critical reading by a human, other aspects can be automatically stress-tested in R.
MotivationAfter a dynamic exercise has been developed, thorough testing is recommended before administering the exercise in a student assessment. Therefore, the R/exams package provides the function
To do so, the function takes the exercise and compiles it hundreds or thousands of times. In each of the iterations the correct solution(s) and the run times are stored in a data frame along with all variables (numeric/integer/character of length 1) created by the exercise. This data frame can then be used to examine undesirable behavior of the exercise. More specifically, for some values of the variables the solutions might become extreme in some way (e.g., very small or very large etc.) or single-choice/multiple-choice answers might not be uniformly distributed. In our experience such patterns are not rare in practice and our students are very good at picking them up, leveraging them for solving the exercises in exams. Example: Calculating binomial probabilitiesIn order to exemplify the work flow, let's consider a simple exercise about calculating a certain binomial probability:
In R, the correct answer for this exercise can be computed as In the folllowing, we illustrate typical problems of parameterizing such an exercise: For an exercise with numeric answer we only need to sample the variables
If you want to replicate the illustrations below you can easily download all four exercises from within R. Here, we show how to do so for the R/Markdown ( Numerical exercise: First attemptIn a first attempt we generate the parameters in the exercise as: Let's stress test this exercise: By default this generates 100 random draws from the exercise with seeds from 1 to 100. The seeds are also printed in the R console, seperated by slashes. Therefore, it is easy to reproduce errors that might occur when running the stress test, i.e., just Here, no errors occurred but further examination shows that parameters have clearly been too extreme: The left panel shows the distribution run times which shows that this was fast without any problems. However, the distribution of solutions in the right panel shows that almost all solutions are extremely small. In fact, when accessing the solutions from the object You might have already noticed the source of the problems when we presented the code above: the values for Remark: In addition to the Numerical exercise: Improved versionTo fix the problems detected above, we increase the range for Stress testing this modified exercise yields solutions with a much better spread: Closer inspection shows that solutions can still become rather small: For fine tuning we might be interested in finding out when the threshold of 5 percent probability is exceeded, depending on the variables So maybe we could increase the minimum for Single-choice exercise: First attemptBuilding on the numeric example above we now move on to set up a corresponding single-choice exercise. This is supported by more Additionally, the code makes sure that we really do obtain five distinct numbers. Even if the probability of two distractors coinciding is very small, it might occur eventually. Finally, because the correct solution is always the first element in So we are quite hopeful that our exercise will do ok in the stress test: The runtimes are still ok (indicating that the And indeed Single-choice exercise: Improved versionHere, we continue to improve the exercise by randomly selecting only one of the two typical errors. Then, sometimes the error is smaller and sometimes larger than the correct solution. Moreover, we leverage the function Again, this is run in a
Finally, the list of possible answers automatically gets LaTeX math markup The corresponding stress test looks sataisfactory now: Of course, there is still at least one number that is either larger or smaller than the correct solution so that the correct solution takes rank 1 or 5 less often than ranks 2 to 4. But this seems to be a reasonable compromise. SummaryWhen drawing many random versions from a certain exercise template, it is essential to thoroughly check that exercise before using it in real exams. Most importantly, of course, the authors should make sure that the story and setup is sensible, question and solution are carefully formulated, etc. But then the technical aspects should be checked as well. This should include checking whether the exercise can be rendered correctly into PDF via The problems in this tutorial could be detected with Note that in our To leave a comment for the author, please follow the link and comment on their blog: R/exams. R-bloggers.com offers daily e-mail updates about R news and tutorials on topics such as: Data science, Big Data, R jobs, visualization (ggplot2, Boxplots, maps, animation), programming (RStudio, Sweave, LaTeX, SQL, Eclipse, git, hadoop, Web Scraping) statistics (regression, PCA, time series, trading) and more... This posting includes an audio/video/photo media file: Download Now | |||||||||||||||||||||
| Posted: 08 Jul 2019 05:00 PM PDT (This article was first published on R Views, and kindly contributed to R-bloggers) Welcome to a mid-summer edition of Reproducible Finance with R. Today, we'll explore the dividend histories of some stocks in the S&P 500. By way of history for all you young tech IPO and crypto investors out there: way back, a long time ago in the dark ages, companies used to take pains to generate free cash flow and then return some of that free cash to investors in the form of dividends. That hasn't been very popular in the last 15 years, but now that it looks increasing likely that interest rates will never, ever, ever rise to "normal" levels again, dividend yields from those dinosaur companies should be an attractive source of cash for a while. Let's load up our packages. We are going to source our data from We want to Let's arrange the Let's import our data from Let's take a look at the most recent dividend paid by each company, to search for any weird outliers. We don't want any 0-dollar dividend dates, so we
A $500 dividend from Google back in 2014? A little, um, internet search engine'ing reveals that Google had a stock split in 2014 and issued that split as a dividend. That's not quite what we want to capture today – in fact, we're pretty much going to ignore splits and special dividends. For now, let's adjust our filter to
Note, this is the absolute cash dividend payout. The dividend yield, the total annual cash dividend divided by the share price, might be more meaningful to us. To get the total annual yield, we want to sum the total dividends in 2018 and divide by the closing price at, say, the first dividend date. To get total dividends in a year, we first create a year column with
Let's nitpick this visualization and take issue with the fact that some of the labels are overlapping – just the type of small error that drives our end audience crazy. It is also a good opportunity to explore the
We have a decent snapshot of these companies' dividends in 2018. Let's dig into their histories a bit. If you're into this kind of thing, you might have heard of the Dividend Aristocrats or Dividend Kings or some other nomenclature that indicates a quality dividend-paying company. The core of these classifications is the consistency with which companies increase their dividend payouts, because those annual increases indicate a company that has strong free cash flow and believes that shareholder value is tied to the dividend. For examples of companies which don't fit this description, check out every tech company that has IPO'd in the last decade. (Just kidding. Actually, I'm not kidding but now I guess I'm obligated to do a follow-up post on every tech company that has IPO'd in the last decade so we can dig into their free cash flow – stay tuned!) Back to dividend consistency, we're interested in how each company has behaved each year, and specifically scrutinizing whether the company increased its dividend from the previous year. This is one of those fascinating areas of exploration that is quite easy to conceptualize and even describe in plain English, but turns out to require (at least for me) quite a bit of thought and white-boarding (I mean Goggling and Stack Overflow snooping) to implement with code. In English, we want to calculate the total dividend payout for each company each year, then count how many years in a row the company has increased that dividend consecutively up to today. Let's get to it with some code. We'll start with a very similar code flow as above, except we want the whole history, no filtering to just 2018. Let's also Let's save that as an object called The data now looks like we're ready to get to work. But have a quick peek at the AAPL data above. Notice anything weird? AAPL paid a dividend of Now we want to find years of consecutive increase, up to present day. We can see that XOM has a nice history of increasing its dividend. For my brain, it's easier to work through the next steps if we put 2018 as the first observation. Let's use Here I sliced off the first 10 rows of each group, so I could glance with my human eyes and see if anything looked weird or plain wrong. Try filtering by Looks like a solid history going back to 2003. In 2004 Now we want to start in 2018 (this year) and count the number of consecutive dividend increases, or 1's in the Let's take it piece-by-painstaking-piece. If we just If we use The magic comes from Now let's order our data by those years of increase, so that the company with the longest consecutive years of increase is at the top. And the winner is….
Before we close, for the curious, I did run this separately on all 505 members of the S&P 500. It took about five minutes to pull the data from
Aren't you just dying to know which ticker is that really high bar at the extreme right? It's A couple of plugs before we close: If you like this sort of thing check out my book, Reproducible Finance with R. I'm also going to be posting weekly code snippets on linkedin, connect with me there if you're keen for some weekly R finance stuff. Happy coding!
To leave a comment for the author, please follow the link and comment on their blog: R Views. R-bloggers.com offers daily e-mail updates about R news and tutorials on topics such as: Data science, Big Data, R jobs, visualization (ggplot2, Boxplots, maps, animation), programming (RStudio, Sweave, LaTeX, SQL, Eclipse, git, hadoop, Web Scraping) statistics (regression, PCA, time series, trading) and more... This posting includes an audio/video/photo media file: Download Now | |||||||||||||||||||||
| Tableau – Creating a Waffle Chart Posted: 07 Jul 2019 05:00 PM PDT (This article was first published on Posts on Anything Data, and kindly contributed to R-bloggers) Waffles are for BreakfastIt's been a long time since my last update and I've decided to start with Tableau, of all topics! Although open source advocates do not look kindly upon Tableau, I find myself using it frequently and relearning all the stuff I can do in R. For my series of "how-to's" regarding Tableau, I'd like to start with posting about how to make a waffle chart in Tableau. Without further ado: What's a Waffle Chart?   There are a gazillion ways to depict an amount. Bar charts and pie charts are common, maybe you've even seen an area/tree chart. But variety is the spice of life – repeatedly looking at bar charts becomes tiring. Enter the waffle chart – It has the appearance of a waffle iron. It's basically a grid of squares (or any other mark). Squares can be colored to represent an amount. In Tableau, a waffle chart is NOT one of the off-the-shelf charts that you can make by clicking the "Show Me" panel. If you want to create one, you'll need to be, err, creative. Setting UpFor this tutorial I will use the famous iris dataset. You can find it anywhere, but it is primarily hosted at https://archive.ics.uci.edu/ml/datasets/iris. Download it and get it into Tableau. Important – Make sure that your spreadsheet software creates an "ID" variable for each row (a row number for each row), and read it as a dimension. Some things to figure out:
Of course, there's always more than one way to do things in Tableau. I'll show my unique way of building a waffle chart which serves my data needs quite well. We start by creating an index. To do so, all you have to do is make a calculated field with INDEX() in it.   Next up, use INDEX to partition the whole dataset into 10 rows to build out the x-axis. So let's figure out how many rows are in a "tenth" of the data – divide the maximum value of the index by 10 or multiply by 0.1. Note it is important to use ROUND to square off the value. If not, you will have rows of unequal lengths later. To make the X-axis from here, I simply assign values based off of where their index stands in relation to each tenth percentile.  At this point, you place your newly created X variable on the rows shelf and choose to "compute using ____" (your dimension/row-level ID variable), and doing so you can see a neat little row of marks as such:  Now we need to expand this single row into 10 Y-values. If you guessed that we will now partition the X-values into 10 different rows, you are right. To do this I multiply the index value by 100 and divide by the maximum index value. This is exactly what it sounds like – a percentage. Note that rounding the values to the nearest integer is very important. Without doing so we won't achieve a neatly separated 10 x 10 grid but a continuous line of marks instead.  The result is not a beautiful 10×10 grid, but a beautiful 11 x 11 grid.  This is because of the rounding we did earlier – for each tenth percentile partiion of X, the bottom portion of the Y values were closer to 0 than 1, which means they were rounded down. Other tutorials may have a workaround, however, they seem to employ other means which require their own unique workarounds as well. At the end of the day, you're probably going to need a workaround. Here is mine: Simply fold the 0 and 1 Y values into each other with another calculated field, as such.  And we get ====> And we get ====> Voila! Now we have our "template" for a waffle chart! Not that it was necessary to fold all the values of our data into the chart. But we can't eat waffles yet – we need to assign color to it first. I suppose I'll color the waffle chart based off of the percentage of Virginica species in the Iris dataset. Notice that I need to use fixed LOD calculations for this. Since the "View" influences calculations, it is best to work with an LOD which will compute the calculation first in spite of whatever is happening in the view.   Now we are ready to assign a color using the percentage variable. The logic is profoundly simple once you figure out the concept. Essentially, you need to tell Tableau to color a mark based off of the values it is less than. That means, multiplying X values by 10, Y values by 1, and starting by looking at the previous row. This calculation handles all possibilities, except of course doing partial squares. 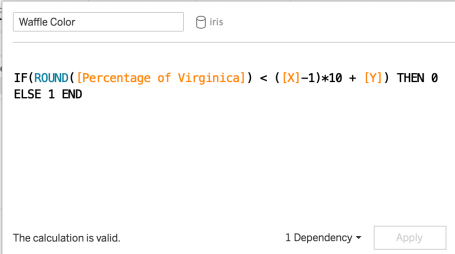 Presto! Now we can finalize the beautiful waffle chart!  33% of the flowers in the iris dataset belong to the virginica species. The End – Enjoy your Waffles!To leave a comment for the author, please follow the link and comment on their blog: Posts on Anything Data. R-bloggers.com offers daily e-mail updates about R news and tutorials on topics such as: Data science, Big Data, R jobs, visualization (ggplot2, Boxplots, maps, animation), programming (RStudio, Sweave, LaTeX, SQL, Eclipse, git, hadoop, Web Scraping) statistics (regression, PCA, time series, trading) and more... This posting includes an audio/video/photo media file: Download Now |
| You are subscribed to email updates from R-bloggers. To stop receiving these emails, you may unsubscribe now. | Email delivery powered by Google |
| Google, 1600 Amphitheatre Parkway, Mountain View, CA 94043, United States | |




 in this blog post. Including now.
in this blog post. Including now.



















Comments
Post a Comment