[R-bloggers] Forget about Excel, Use these R Shiny Packages Instead (and 2 more aRticles) | |
- Forget about Excel, Use these R Shiny Packages Instead
- Analyzing a binary outcome arising out of within-cluster, pair-matched randomization
- Spatial regression in R part 1: spaMM vs glmmTMB
| Forget about Excel, Use these R Shiny Packages Instead Posted: 03 Sep 2019 12:58 AM PDT [This article was first published on r – Appsilon Data Science | End to End Data Science Solutions, and kindly contributed to R-bloggers]. (You can report issue about the content on this page here) Want to share your content on R-bloggers? click here if you have a blog, or here if you don't.  tl; drTransferring your Excel sheet to a Shiny app can be the easiest way to create an enterprise ready dashboard. In this post, I present 6 Shiny alternatives for the table-like data that Excel users love. IntroExcel has its limitations regarding advanced statistics and calculations, quality and version control, user experience and scalability. Switching to a more sophisticated data analysis tool or a dashboard is often an answer. Transferring your Excel sheet to a Shiny app can be the easiest way to create an enterprise ready dashboard. Take a look at Filip Stachura's article "Excel Is Obsolete" which addresses, from the architecture point of view, when to stick with Excel and when it is time for a change In this blogpost we'll focus on the functionalities that may be implemented in a Shiny app. You're probably aware of Shiny's cool interactive plot and charts features that are well ahead of what you can do in Excel. What may still prevent you from switching from Excel to a more advanced Shiny dashboard is the fear of missing the beloved Excel functionalities to work with table-like data. Don't worry! It is super easy to implement and extend them using Shiny. The most commonly used table widgets in Shiny are DT and rhandsontable. Let's take a deep dive into their features but also look at some other packages strictly dedicated to help with popular spreadsheets tasks. 1. Editable tablesA basic reason for using a spreadsheet is the simplicity of data manipulation. Displaying data is not always enough. Content may require spell checking, fixing, adding rows or columns. A similar solution is available in excelR. The package is worth testing as it contains many interesting solutions such as radio selection inside table and the multiple well-known Excel functions, like SUM presented below. The package also allows users to easily manipulate cells with actions like resizing, merging or switching row/column positions. Nested headers are also useful as they can organize your data. Implementing the solution in Shiny is easy and intuitive with well-known render/output dedicated functions. There are two downsides with excelR at the moment — cloning formulas between columns, and calculation approximations, which do not work as one would expect. The DT package has a lot of great features and is a great option when heavy data editing is not the main goal. And as you can see in the gif below, tables implemented with DT look really nice. It has less functionality than rhandsontable though, basically just allowing the user to replace the values inside cells when double-clicked, without validating the values typed in. Some columns may be restricted to be read-only.  Source: https://rstudio.github.io/DT/ A possible workaround for DT's limited editing functionality is a more advanced DTedit package. It comes with a pleasing interface (modal dialog) for editing single table rows as well as buttons to add, delete or copy data. The package is currently only available on GitHub, but we will keep our fingers crossed for its expansion and increase in popularity.  Source: https://github.com/jbryer/DTedit 2. Conditional formattingConditional formatting is a super useful tool for getting a quick overview when dealing with tons of values. Both rhandsontable and DT allow users to format cells according to its values. If your highest priority for your application is beautiful data presentation, then the package formattable is worth checking out. The formatting interface is more user-friendly than in rhandsontable and it is based on R functions, not pure javascript code. Besides working on tables, it also contains functionality of formatting R vectors which might be useful when presenting results of pure R analysis.
3. Sorting and filteringSorting and filtering are also crucial when examining a huge dataset. In the rhandsontable package, sorting columns can be enabled by a single parameter, however filtering is not implemented inside the feature and may require adding some extra Shiny components. On the other hand, in DT, column sorting is available by default as well as global search. Enabling column filtering is as easy as adding a single parameter (filter = top/bottom, depending on where the filters should be placed).  Source: https://rstudio.github.io/DT/ 4. Drag & drop pivot tablesExcel users love pivot tables. Allowing the users to create their own stories based on data is an excellent feature – sometimes the valuable info is only generated when looking at the data from the right angle. For a plug-and-play pivot table we recommend using the rpivotTable package. As you can see in the gif, it is super easy to produce tables and manipulate the aggregation variables with drag & drop. You can also filter by specific values and/or choose which variable should be calculated based on the selected function – like presented below sum and sum as percentage of total. Quickly switching from table to different types of (interactive!) charts is a great bonus. If you would like to combine pivoting with other features, a combination of shinydnd and DT Custom Table Containers as well as data manipulation is needed. Nevertheless, the results can be amazing. Maybe we'll get into that in a future post. 5. Reacting on selectionThe usual scenario in dashboards applications is reacting to user selection and continuing to work on a selected element. When it needs to be a key feature of the application, then the DT package is a great choice. It is easy to implement logic for reacting to user cell/row/column selection. The sky's the limit really! Options range from custom edit data tools to going deep into nested tables. Or as presented in the gif below, you see the graph dynamically reacts to user selection in the table. 6. Expandable rows…is an extra nice feature that allows you to hide (in an elegant way) additional information and bring the crucial part to the top. This is another feature that does not exist in spreadsheets. Expandable rows is also useful in presenting database-like structures with one-to-many relations. It requires a little javascript magic in DT for now, but the various examples (including this one) are easy to follow. ConclusionYou may have felt that if you switched from Excel to Shiny, you would be limited in the table data feature set. I hope you can see by now that Shiny offers a comparable feature set to Excel as well as exciting new possibilities! You can reach me on Twitter @DubelMarcin. Article Forget about Excel, Use these R Shiny Packages Instead comes from Appsilon Data Science | End to End Data Science Solutions. To leave a comment for the author, please follow the link and comment on their blog: r – Appsilon Data Science | End to End Data Science Solutions. R-bloggers.com offers daily e-mail updates about R news and tutorials about learning R and many other topics. Click here if you're looking to post or find an R/data-science job. Want to share your content on R-bloggers? click here if you have a blog, or here if you don't. This posting includes an audio/video/photo media file: Download Now |
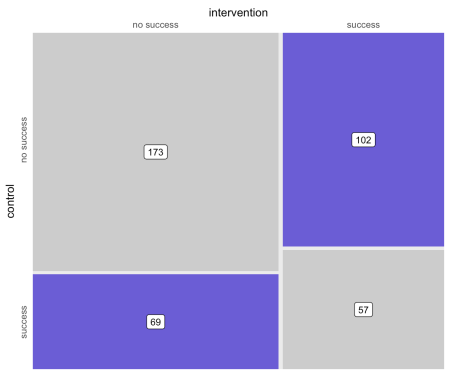
| Analyzing a binary outcome arising out of within-cluster, pair-matched randomization Posted: 02 Sep 2019 05:00 PM PDT [This article was first published on ouR data generation, and kindly contributed to R-bloggers]. (You can report issue about the content on this page here) Want to share your content on R-bloggers? click here if you have a blog, or here if you don't. A key motivating factor for the The study designThe original post has the details about the design and matching algorithm (and code). The randomization is taking place at 20 primary care clinics, and patients within these clinics are matched based on important characteristics before randomization occurs. There is little or no risk that patients in the control arm will be "contaminated" or affected by the intervention that is taking place, which will minimize the effects of clustering. However, we may not want to ignore the clustering altogether. Possible analytic solutionsGiven that the primary outcome is binary, one reasonable procedure to assess whether or not the intervention is effective is McNemar's test, which is typically used for paired dichotomous data. However, this approach has two limitations. First, McNemar's test does not take into account the clustered nature of the data. Second, the test is just that, a test; it does not provide an estimate of effect size (and the associated confidence interval). So, in addition to McNemar's test, I considered four additional analytic approaches to assess the effect of the intervention: (1) Durkalski's extension of McNemar's test to account for clustering, (2) conditional logistic regression, which takes into account stratification and matching, (3) standard logistic regression with specific adjustment for the three matching variables, and (4) mixed effects logistic regression with matching covariate adjustment and a clinic-level random intercept. (In the mixed effects model, I assume the treatment effect does not vary by site, since I have also assumed that the intervention is delivered in a consistent manner across the sites. These may or may not be reasonable assumptions.) While I was interested to see how the two tests (McNemar and the extension) performed, my primary goal was to see if any of the regression models was superior. In order to do this, I wanted to compare the methods in a scenario without any intervention effect, and in another scenario where there was an effect. I was interested in comparing bias, error rates, and variance estimates. Data generationThe data generation process parallels the earlier post. The treatment assignment is made in the context of the matching process, which I am not showing this time around. Note that in this initial example, the outcome Based on the outcomes of each individual, each pair can be assigned to a particular category that describes the outcomes. Either both fail, both succeed, or one fails and the other succeeds. These category counts can be represented in a \(2 \times 2\) contingency table. The counts are the number of pairs in each of the four possible pairwise outcomes. For example, there were 173 pairs where the outcome was determined to be unsuccessful for both intervention and control arms. Here is a figure that depicts the \(2 \times 2\) matrix, providing a visualization of how the treatment and control group outcomes compare. (The code is in the addendum in case anyone wants to see the lengths I took to make this simple graphic.)
McNemar's testMcNemar's test requires the data to be in table format, and the test really only takes into consideration the cells which represent disagreement between treatment arms. In terms of the matrix above, this would be the lower left and upper right quadrants. Based on the p-value = 0.01, we would reject the null hypothesis that the intervention has no effect. Durkalski extension of McNemar's testDurkalski's test also requires the data to be in tabular form, though there essentially needs to be a table for each cluster. The Again, the p-value, though larger, leads us to reject the null. Conditional logistic regressionConditional logistic regression is conditional on the pair. Since the pair is similar with respect to the matching variables, no further adjustment (beyond specifying the strata) is necessary.
Logistic regression with matching covariates adjustmentUsing logistic regression should in theory provide a reasonable estimate of the treatment effect, though given that there is clustering, I wouldn't expect the standard error estimates to be correct. Although we are not specifically modeling the matching, by including covariates used in the matching, we are effectively estimating a model that is conditional on the pair.
Generalized mixed effects model with matching covariates adjustmentThe mixed effects model merely improves on the logistic regression model by ensuring that any clustering effects are reflected in the estimates.
Comparing the analytic approachesTo compare the methods, I generated 1000 data sets under each scenario. As I mentioned, I wanted to conduct the comparison under two scenarios. The first when there is no intervention effect, and the second with an effect (I will use the effect size used to generate the first data set. I'll start with no intervention effect. In this case, the outcome definition sets the true parameter of Using the updated definition, I generate 1000 datasets, and for each one, I apply the five analytic approaches. The results from each iteration are stored in a large list. (The code for the iterative process is shown in the addendum below.) As an example, here are the contents from the 711th iteration: Summary statisticsTo compare the five methods, I am first looking at the proportion of iterations where the p-value is less then 0.05, in which case we would reject the the null hypothesis. (In the case where the null is true, the proportion is the Type 1 error rate; when there is truly an effect, then the proportion is the power.) I am less interested in the hypothesis test than the bias and standard errors, but the first two methods only provide a p-value, so that is all we can assess them on. Next, I calculate the bias, which is the average effect estimate minus the true effect. And finally, I evaluate the standard errors by looking at the estimated standard error as well as the observed standard error (which is the standard deviation of the point estimates). In this first case, where the true underlying effect size is 0, the Type 1 error rate should be 0.05. The Durkalski test, the conditional logistical regression, and the mixed effects model are below that level but closer than the other two methods. All three models provide unbiased point estimates, but the standard logistic regression (glm) underestimates the standard errors. The results from the conditional logistic regression and the mixed effects model are quite close across the board. Here are the summary statistics for a data set with an intervention effect of 0.45. The results are consistent with the "no effect" simulations, except that the standard linear regression model exhibits some bias. In reality, this is not necessarily bias, but a different estimand. The model that ignores clustering is a marginal model (with respect to the site), whereas the conditional logistic regression and mixed effects models are conditional on the site. (I've described this phenomenon here and here.) We are interested in the conditional effect here, so that argues for the conditional models. The conditional logistic regression and the mixed effects model yielded similar estimates, though the mixed effects model had slightly higher power, which is the reason I opted to use this approach at the end of the day. In this last case, the true underlying data generating process still includes an intervention effect but no clustering. In this scenario, all of the analytic yield similar estimates. However, since there is no guarantee that clustering is not a factor, the mixed effects model will still be the preferred approach. The DREAM Initiative is supported by the National Institutes of Health National Institute of Diabetes and Digestive and Kidney Diseases R01DK11048. The views expressed are those of the author and do not necessarily represent the official position of the funding organizations.
Addendum: multiple datasets and model estimates
Code to generate figureTo leave a comment for the author, please follow the link and comment on their blog: ouR data generation. R-bloggers.com offers daily e-mail updates about R news and tutorials about learning R and many other topics. Click here if you're looking to post or find an R/data-science job. Want to share your content on R-bloggers? click here if you have a blog, or here if you don't. This posting includes an audio/video/photo media file: Download Now |
| Spatial regression in R part 1: spaMM vs glmmTMB Posted: 02 Sep 2019 08:17 AM PDT [This article was first published on R Programming – DataScience+, and kindly contributed to R-bloggers]. (You can report issue about the content on this page here) Want to share your content on R-bloggers? click here if you have a blog, or here if you don't. CategoryTagsMany datasets these days are collected at different locations over space which may generate spatial dependence. Spatial dependence (observation close together are more correlated than those further apart) violate the assumption of independence of the residuals in regression models and require the use of a special class of models to draw the valid inference. The first thing to realize is that spatial data come in very different forms: areal data (murder rate per county), point pattern (trees in forest – random sampling locations) or point referenced data (soil carbon content – non random sampling locations), and all of these forms have specific models and R packages such as spatialreg for areal data or spatstat for point pattern. In this blog post I will introduce how to perform, validate and interpret spatial regression models fitted in R on point referenced data using Maximum Likelihood with two different packages: (i) spaMM and (ii) glmmTMB. If you, reader, are aware of other packages out there to fit these models do let me know and I'll be happy to include it in this post. Do I need a spatial model?Before plugging into new model complexity, the first question to ask is: "do my dataset require me to take spatial dependence into account?". The basic steps to answer this question are:
Let's look at this with a first simulated example. In a research project we are interested in understanding the link between tree height and temperature, to achieve this we recorded both parameters in 100 different forests and we want to use regression models. Step 0: data simulationStep 1: fit a non-spatial modelStep 2: test for spatial autocorrelation in the residualsVisually checking model residuals show that there seems to be little spatial dependency there. p-values fans can also rely on formal test (like Moran's I): No evidence of spatial Autocorrelation. We can go to Step 3a. Step 3aThis little toy example showed that even if there are a spatial pattern in the data this does not mean that spatial regression models should be used. In some cases spatial patterns in the response variable are generated by spatial patterns present in the covariates, such as temperature gradient, elevation … Once we take into account the effect of these covariates spatial patterns in the response variable disappear. So before starting to fit a spatial model, one should check that such complexity is warranted by the data. Fitting a spatial regression modelThe basic model structure that we will consider in this post is: \[ y_i \sim \mathcal{N}(\mu_i, \sigma) \] i index the different observations, y is the response variable (tree height …), \(\mu\) is the linear predictor and \(\sigma\) is the residual standard deviation. The linear predictor is defined as follow: \[ \mu_i = X_i\beta + u(s_i) \] \[ u(s_i) \sim \mathcal{MVN}(0, F(\theta_1, …, \theta_n)) \] This model basically translate the expectation that closer observations should be more correlated, the strength of the spatial signal and its decay will have to be estimated from the data and we will explore here two packages to do so: spaMM and glmmTMB. I will show some code to fit the models, interpret the outputs, derive spatial predictions and check model assumptions for all three methods. The datasetspaMMspaMM fits mixed-effect models and allow the inclusion of spatial effect in different forms (Matern, Interpolated Markov Random Fields, CAR / AR1) but also provide interesting other features such as non-gaussian random effects or autocorrelated random coefficient (ie group-specific spatial dependency). spaMM uses a syntax close to the one used in lme4, the main function to fit the model is fitme. We will fit the model structure outlined above to the calcium dataset: There are two main output interesting here: first are the fixed effect (beta) which are the estimated regression parameters (slopes). Then the correlation parameter nu and rho which represent the strength and the speed of decay in the spatial effect, which we can turn into the actual spatial correlation effect by plotting the estimated correlation between two locations against their distance: So basically locations more than 200m away have a correlation below 0.1. Now we can check the model using DHARMa: Looks relatively ok. Now we can predict the effect of elevation and region while controlling for spatial effects: We can also derive prediction at any spatial location provided that we feed in information on the elevation and the region: That's it for spaMM a great, fast and easy way to fit spatial regressions. glmmTMBglmmTMB fits a broad class of GLMM using Template Model Builder. With this package we can fit different covariance structure including spatial Matern. Let's dive right in. The output from glmmTMB should be familiar to frequent users of lme4, first we have some general model information (family, link, formula, AIC …), then we have the estimation of the random effect variance (estimated at 105 here very close to the value from spaMM), and the last table under "Conditional model" display the estimates for the fixed effects. These are again pretty close to those estimated by spaMM. Before going any further let's check the model fitness. By running these lines we get a warning that glmmTMB does not implement yet unconditional predictions (without random effect) is not yet possible in glmmTMB so one may expect (and we do see) some upward going slopes on the right graphs. Not much to do at this stage, maybe in the (near) future with update to glmmTMB this could be solved. We can look at the predicted spatial effect: We find here a similar picture to the one from spaMM, but this was quite a bit more work to get it and we don't seem to have direct estimation of the Matern parameters that were readily available in spaMM. We can now look at the effect of elevation and region (since there is no way to marginalize over the random effects in glmmTMB we have to get the CI by hand): Looks very close to the picture we got from spaMM, which is reassuring. It is a bit more laborious to get estimation of Confidence Intervals for glmmTMB as of know but in th enear future new implementation of the predict method with a "re.form=NA" argument would allow easier derivation of the CIs. Now let's see how to derive spatial predictions: Very similar map to spaMM. ConclusionTime to wrap up what we've seen here. First off, before plunging into spatial regression models you should first check that your covariates do not already take into account the spatial patterns present in your data. Remember that in the implementation of the spatial models discussed here, spatial effects are modelled in a similar fashion to random effects (basically random effect take into account structure in the data be it by design or spatial or temporal structure). So any variation (including spatial) that can be explained by the fixed effects will be taken into account and only remaining variation based on the spatial structure will be effectively going into the estimated spatial effects. Now I presented here two ways to fit similar spatial regression models in R, time to compare a bit their performance and their pros and cons.
In a second part we will explore Bayesian ways to do spatial regression in R with the same dataset, stay tuned for more fun!
Related Post
To leave a comment for the author, please follow the link and comment on their blog: R Programming – DataScience+. R-bloggers.com offers daily e-mail updates about R news and tutorials about learning R and many other topics. Click here if you're looking to post or find an R/data-science job. Want to share your content on R-bloggers? click here if you have a blog, or here if you don't. This posting includes an audio/video/photo media file: Download Now |
| You are subscribed to email updates from R-bloggers. To stop receiving these emails, you may unsubscribe now. | Email delivery powered by Google |
| Google, 1600 Amphitheatre Parkway, Mountain View, CA 94043, United States | |

















[ ] I'm here to testify about the great work Dr Osebor did for me. I have been suffering from (HERPES) disease for the past 5 years and had constant pain, especially in my knees. During the first year,I had faith in God that i would be healed someday.This disease started circulating all over my body and i have been taking treatment from my doctor, few weeks ago I came across a testimony of one lady on the internet testifying about a Man called Dr Osebor on how he cured her from HIV Virus. And she also gave the email address of this man and advise anybody to contact Dr Osebor for help for any kind of sickness that he would be of help, so I emailed him on ( oseborwinbacktemple@gmail.com ) telling him about my (HERPES Virus) he told me not to worry that i was going to be cured!! Well i never believed it,, well after all the procedures and remedy given to me by this man few weeks later i started experiencing changes all over me as Dr Osebor assured me that i will be cured,after some time i went to my doctor to confirmed if i have be finally healed behold it was TRUE, So
ReplyDelete- [ ] friends my advise is if you have such sickness or any other at all you can contact Dr Osebor via email. { oseborwinbacktemple@gmail.com }or call or what sapp him on( +2348073245515 )
- [ ] DR osebor CAN AS WELL CURE THE FOLLOWING DISEASE:-
- [ ] HIV/AIDS
- [ ] HERPES
- [ ] CANCER
- [ ] ALS
- [ ] cancer
- [ ] Diabetes
eye problem etc.